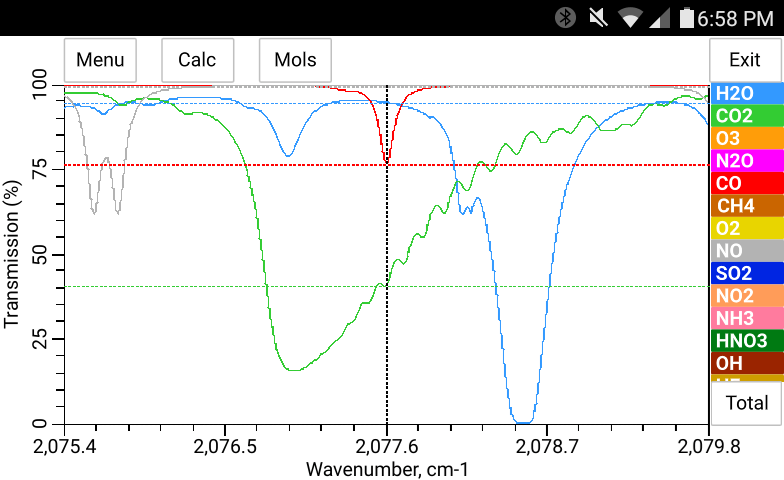
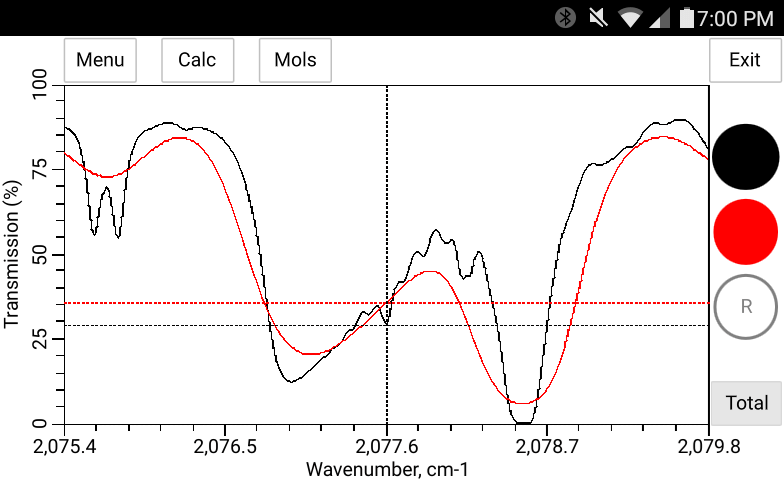
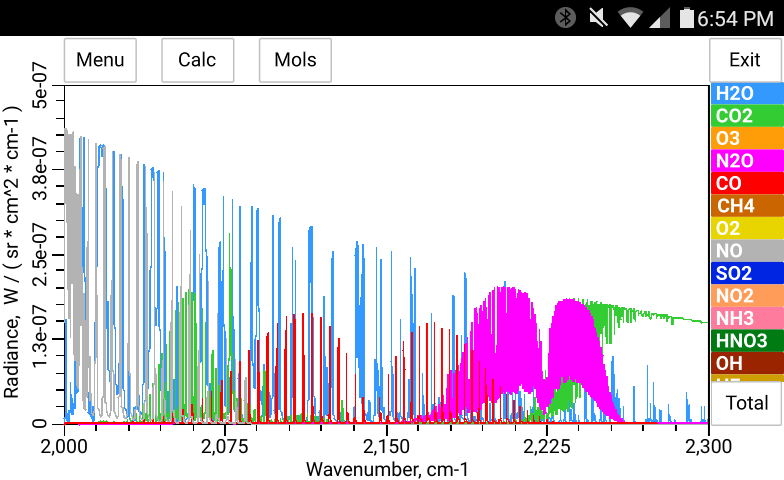
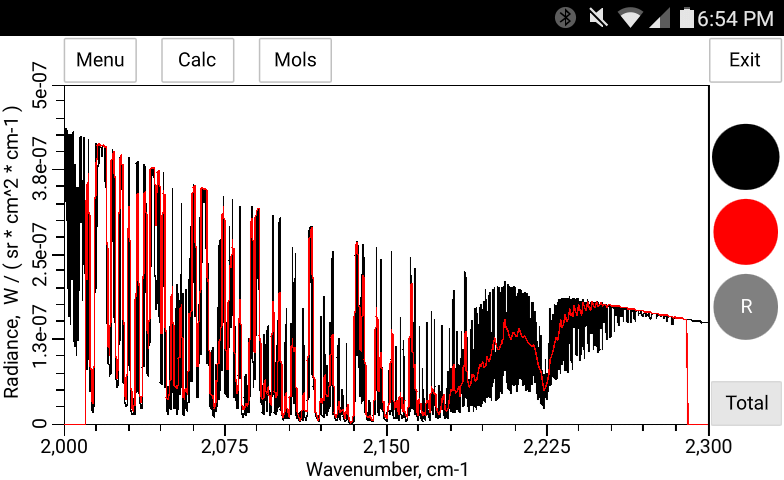
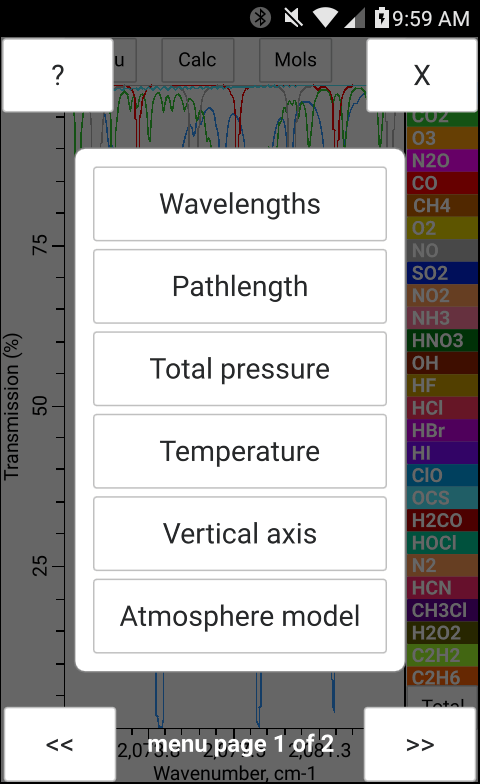
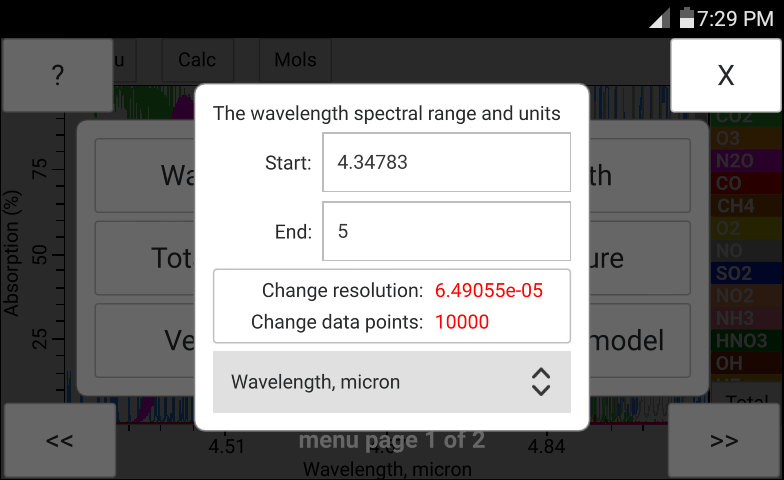
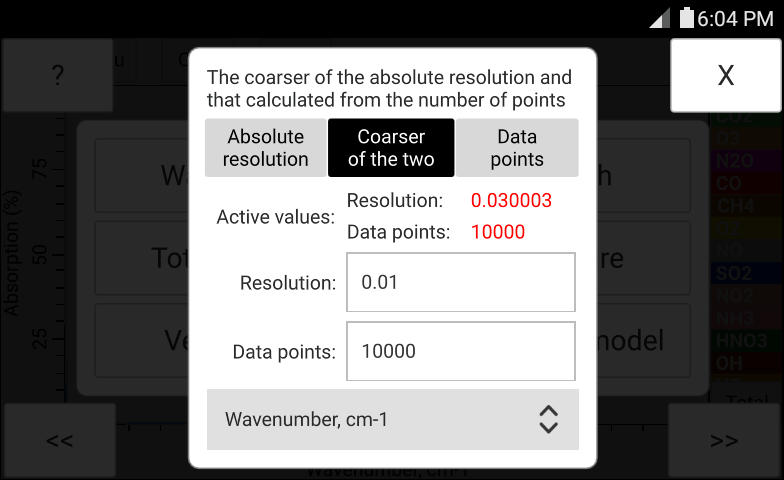
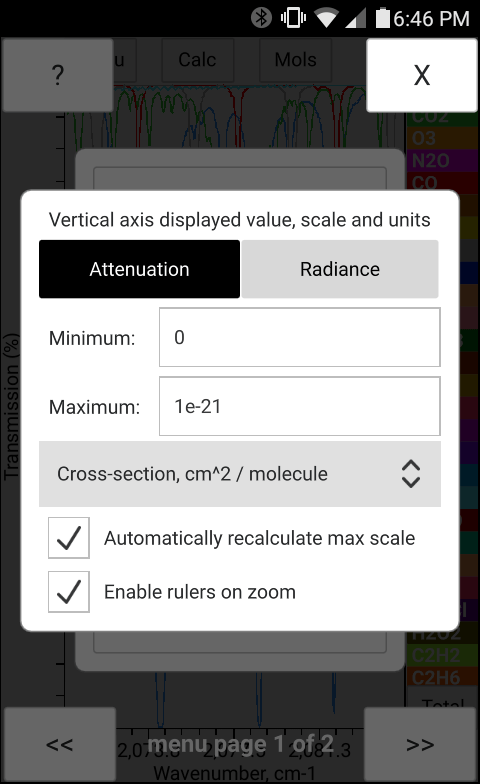
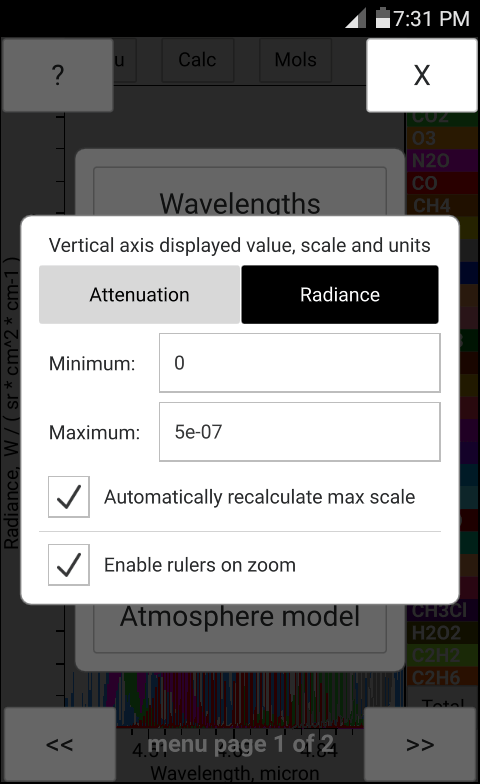
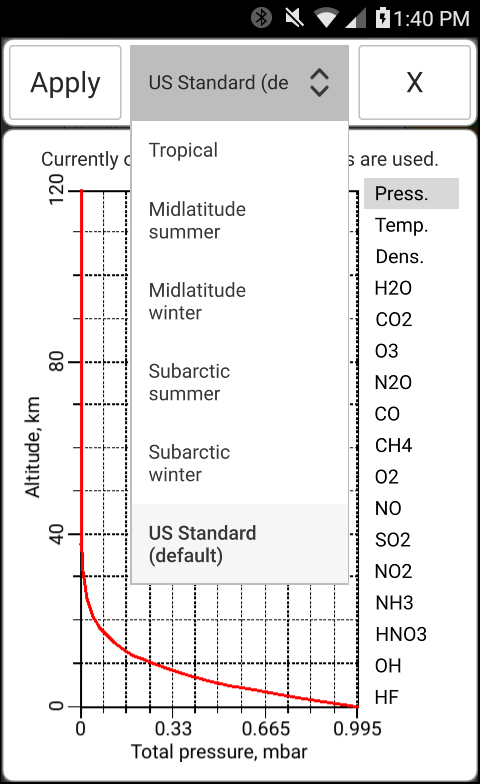
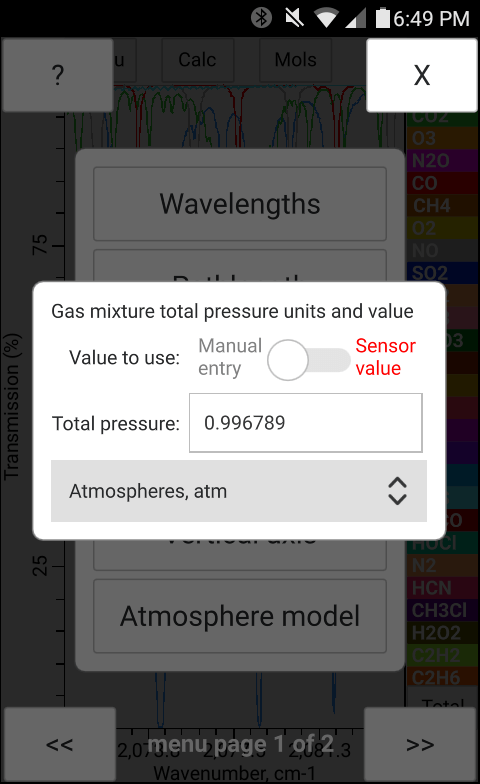
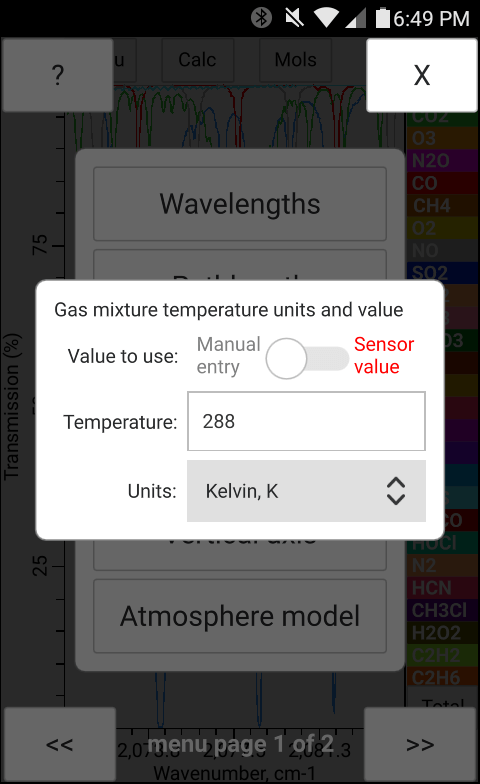
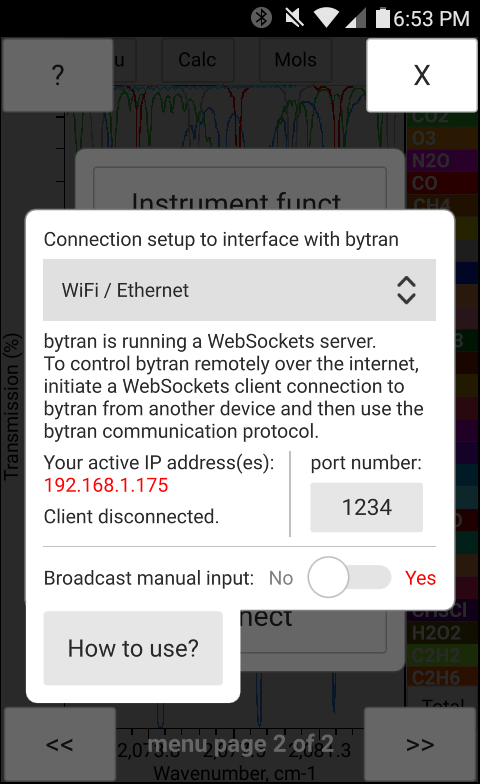
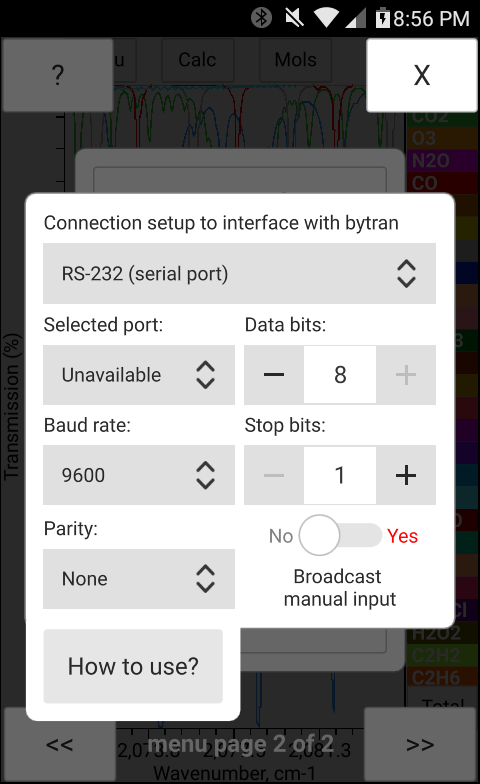
The total pressure, temperature and the concentration of H2O may be entered manually in bytran or else read in from built in or external sensors.
Version 1.3 of bytran supports:
(1) Cell phone built in Temperature and Total pressure sensors under Android,
(2) External bluetooth SensorTag module (Baromter, Thermometer and the Humidity sensors),
(3) Raspberry Pi's Sense HAT add-on board (Baromter, Thermometer and the Humidity sensors).
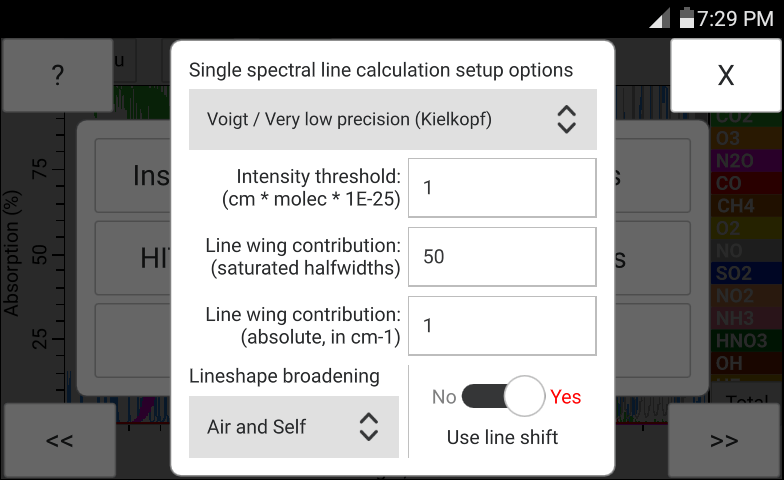
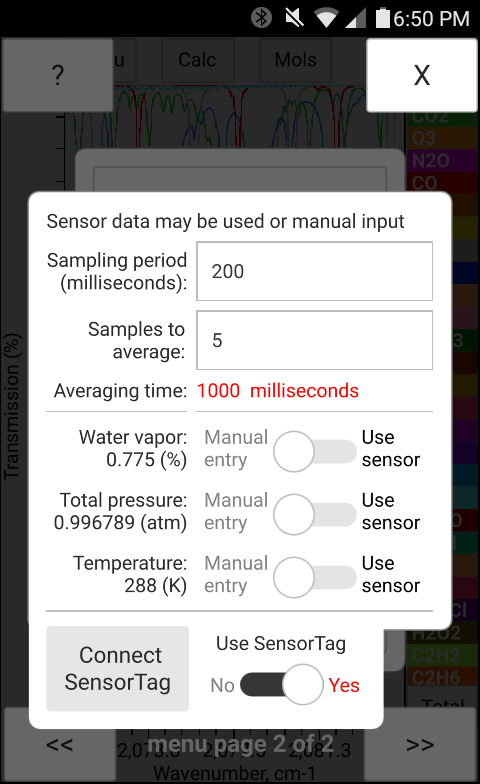
The "Sensors" menu option shown in Figure 17 contains sensor related settings. Among the settings are the "Sampling period" and the "Number of samples to average". The total averaging time calculated as the product of the sampling period and the number of samples to averege is displayed in red. The switches located under the settings menu as shown in Figure 17 which allow switching between the Manual and Sensor input correspond to either built in sensors (if present), or to the SensorTag module (if it is connected and enabled). The bottom switch titled "Use SensorTag module" is responsible for switching between the built in sensors and the input from the SensorTag module. While the built in sensors are detected automatically if present, the SensorTag module needs to be connected over Bluetooth before it can be enabled and used.
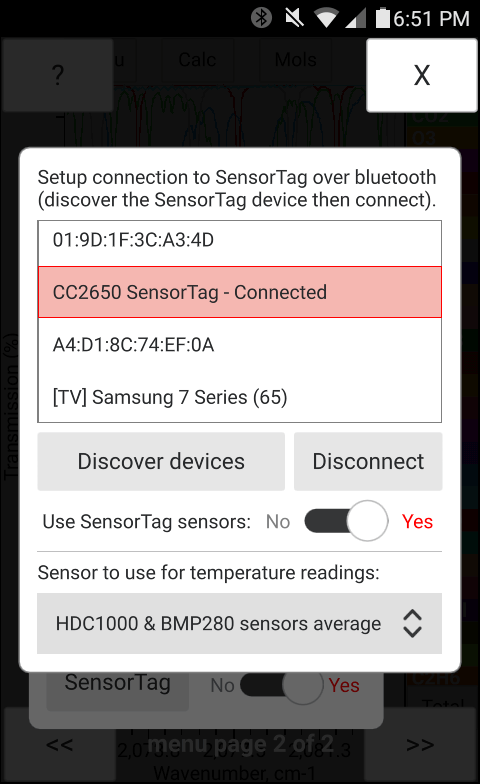
In order to connect the SensorTag module, click on the "Connect SensorTag" button in the "Sensors" window (Figure 17) to open the SensorTag connection window shown in Figure 18 and press the "Discover devices" button after enabling the "Advertizing mode" within the SensorTag module. (The "Advertizing mode" in the SensorTag may be enabled by pressing and holding the smaller size button for a couple of seconds. When the advertizing mode is enabled a green LED in the SensorTag module will start blinking repeatedly.) Once the bluetooth SensorTag discovery is complete in bytran and the list (Figure 18) is populated with the found bluetooth devices, select the SensorTag record in the list and click "Connect". Once the connection is established, the SensorTag record in the list with be updated to state "Connected". The SensorTag has 2 different temperature sensors, it is possilbe to select input from either one of the sensors or their average (the average is used by default). To start using the SensorTag set the "Use SensorTag" switch to "Yes" and enable the desired sensors using switches displayed in the Sensors window (Figure 18).
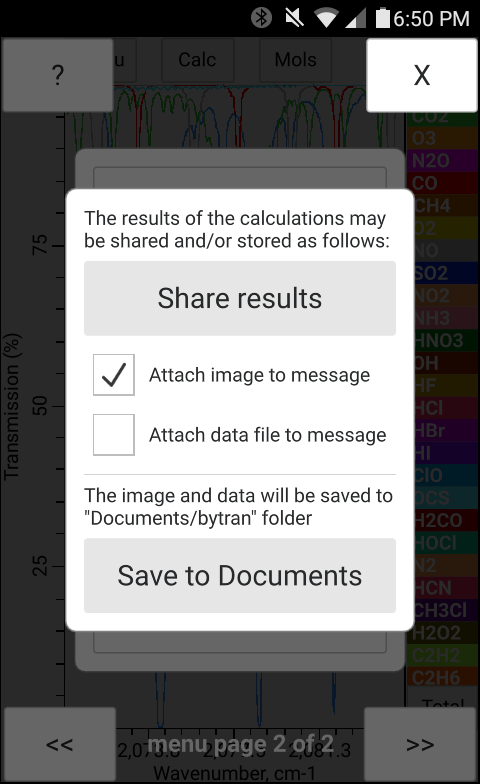
The results of the calculation may be shared under Android and iOS over available communication channels (i.e. email, text messaging etc.). The generated files which may be shared include the (1) spectrum plot image, (2) the datafile with the corresponding numerical data, and the (3) configuration file with the current configuration settings. The user may select to only share the image, the numerical data, or both (see checkbox options in Fig. 18). If the "attach data file to message" is selected, the calculation results data file and the configuration file will be shared. When the results of the calculation are being shared a copy of all shared files is saved into the "Documents/bytran" folder if the operating system has a permission to access the "Documents" folder, otherwise the shared data will be saved into the OS specific application data folder not directly accessible by the user. Under desktop operating systems the data cannot me shared but may be saved to the "Documents/bytran" folder by pressing the "Save to Documents" button shown in Fig. 18 above. Currently bytran does not provide a way to modify the folder where the results of the calculations are stored.